Report for Biomek_20th_2018-06-26
Frank (zhangc518@gmail.com) @ UCSD Center for Epigenomics
Fri Jun 29 20:33:55 2018
Sample info.
| Sample Index (from MSTS) | sample ID (from MSTS) | Label (for QC report) | species | Sequencing ID | Experiment type | Machine | |
|---|---|---|---|---|---|---|---|
| JYH_501 | SAMP-159 | GM19240 | GM19240_3. | Human | JYH_501 | ATAC-seq | NextSeq550 Mid |
| JYH_502 | SAMP-159 | GM19240 | GM19240_2. | Human | JYH_502 | ATAC-seq | NextSeq550 Mid |
| JYH_503 | SAMP-159 | GM19240 | GM19240_1.75. | Human | JYH_503 | ATAC-seq | NextSeq550 Mid |
| JYH_504 | SAMP-159 | GM19240 | GM19240_1.5. | Human | JYH_504 | ATAC-seq | NextSeq550 Mid |
| JYH_505 | SAMP-159 | GM19240 | GM19240_1.25. | Human | JYH_505 | ATAC-seq | NextSeq550 Mid |
| JYH_506 | SAMP-159 | GM19240 | GM19240_1. | Human | JYH_506 | ATAC-seq | NextSeq550 Mid |
| JYH_507 | SAMP-159 | GM19240 | GM19240_0.75. | Human | JYH_507 | ATAC-seq | NextSeq550 Mid |
| JYH_508 | SAMP-159 | GM19240 | GM19240_0.5. | Human | JYH_508 | ATAC-seq | NextSeq550 Mid |
| JYH_509 | Other | HepG2 | HepG2_3. | Human | JYH_509 | ATAC-seq | NextSeq550 Mid |
| JYH_510 | Other | HepG2 | HepG2_2. | Human | JYH_510 | ATAC-seq | NextSeq550 Mid |
| JYH_511 | Other | HepG2 | HepG2_1.75. | Human | JYH_511 | ATAC-seq | NextSeq550 Mid |
| JYH_512 | Other | HepG2 | HepG2_1.5. | Human | JYH_512 | ATAC-seq | NextSeq550 Mid |
| JYH_513 | Other | HepG2 | HepG2_1.25. | Human | JYH_513 | ATAC-seq | NextSeq550 Mid |
| JYH_514 | Other | HepG2 | HepG2_1. | Human | JYH_514 | ATAC-seq | NextSeq550 Mid |
| JYH_515 | Other | HepG2 | HepG2_0.75. | Human | JYH_515 | ATAC-seq | NextSeq550 Mid |
| JYH_516 | Other | HepG2 | HepG2_0.5. | Human | JYH_516 | ATAC-seq | NextSeq550 Mid |
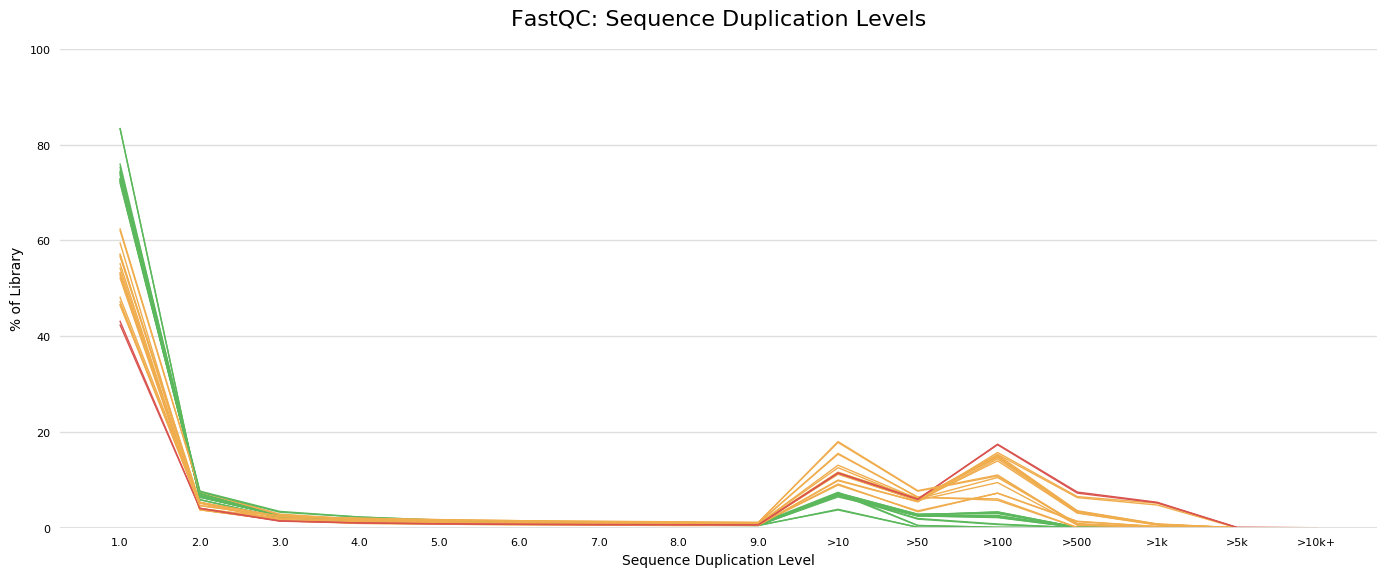
Fastq files
Sequence sources (potential contamination)
Mappability
Reads yeild table
Read yield (%) after each step
Read counts after each step
Mitochondrial reads fraction
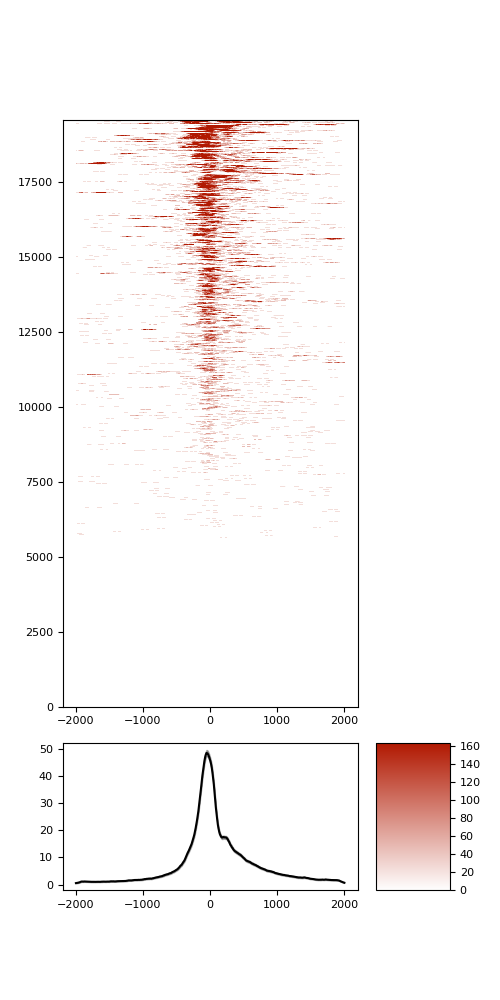
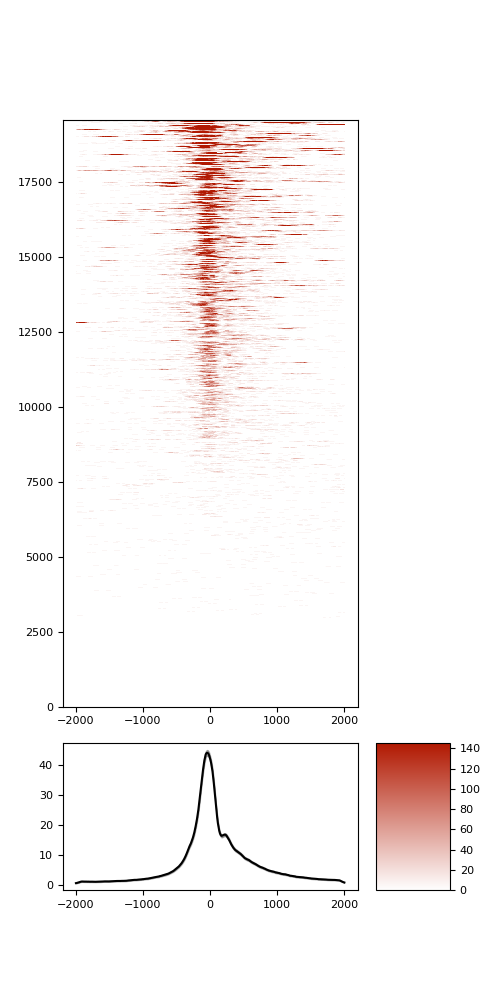
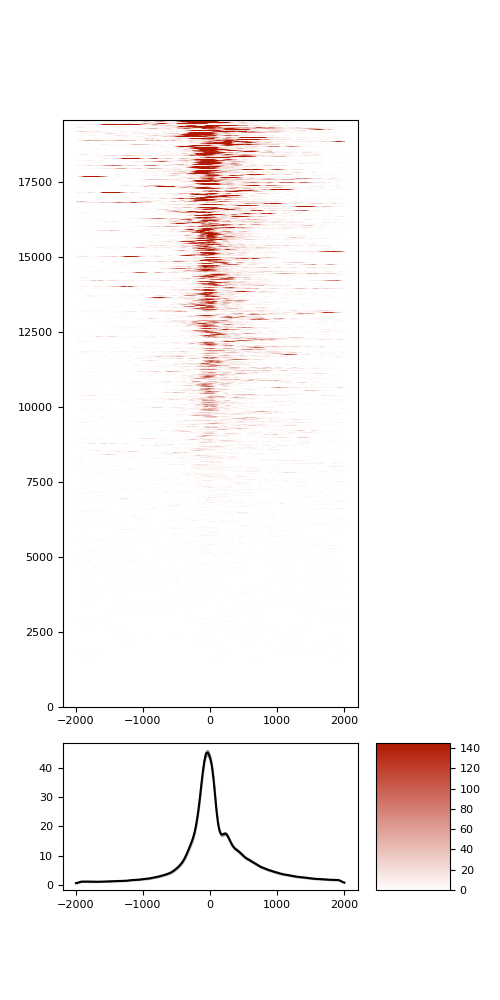
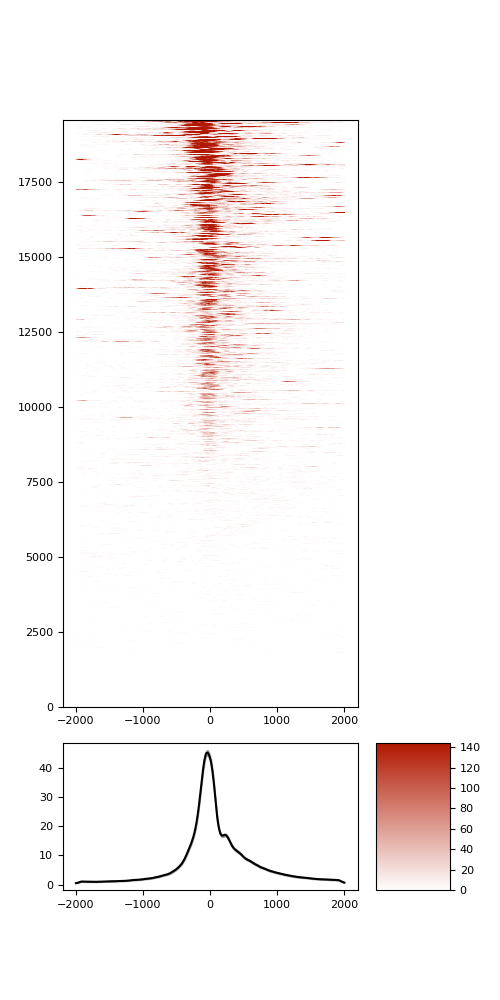
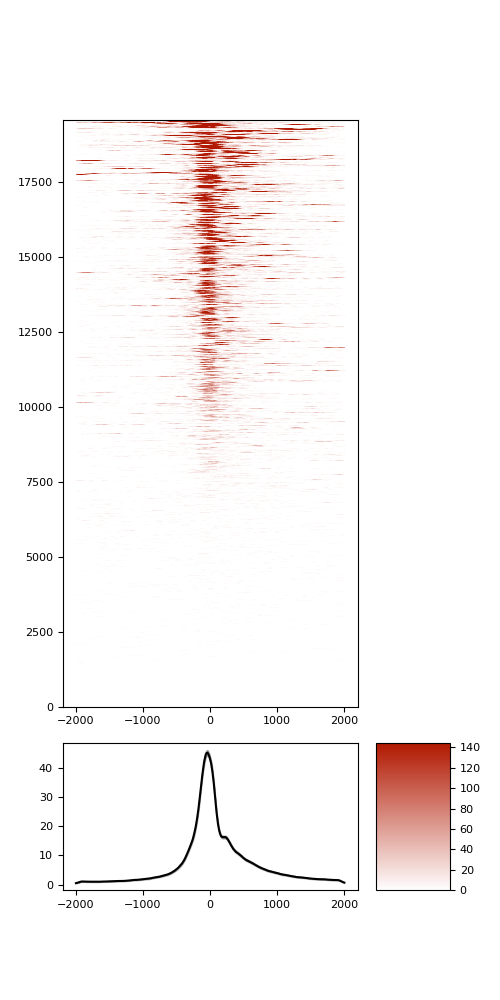
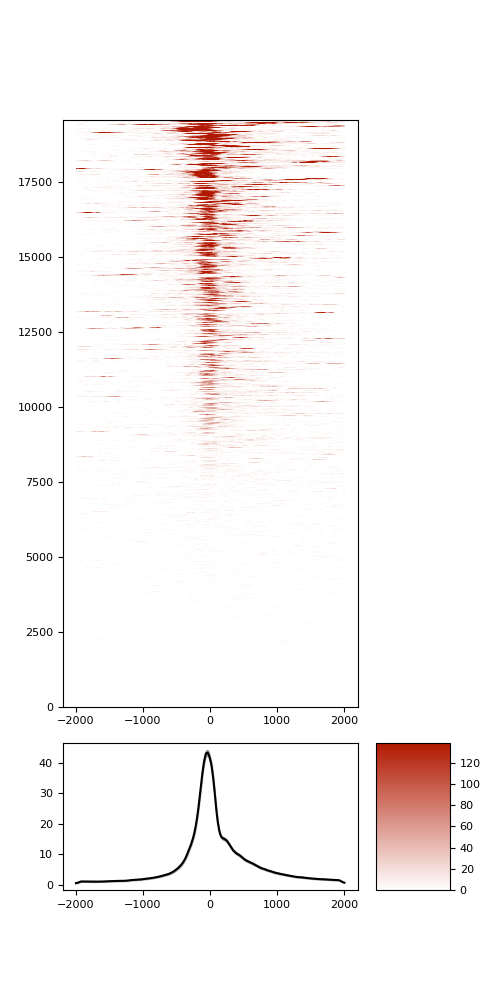
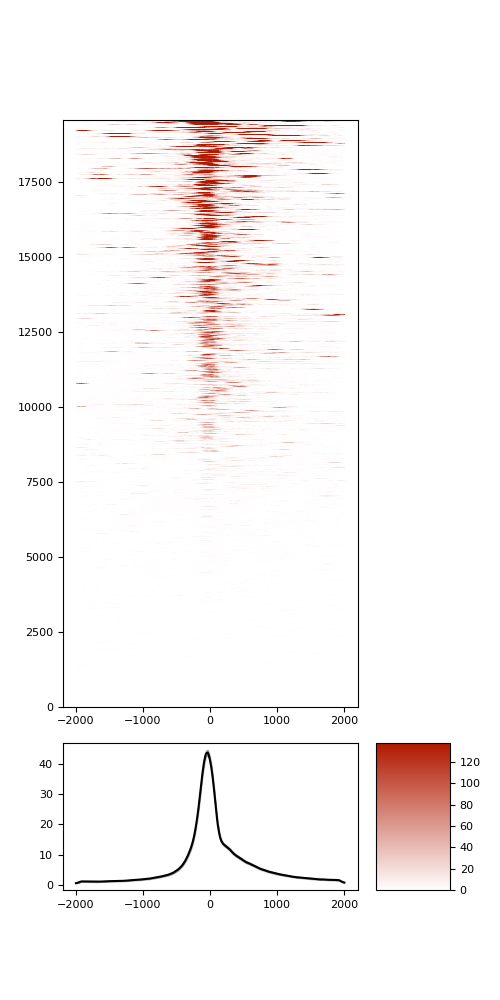
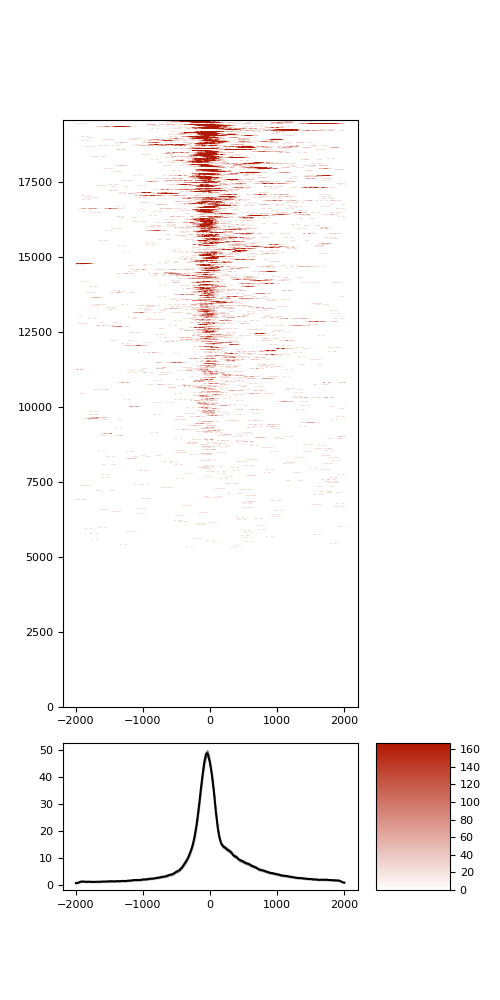
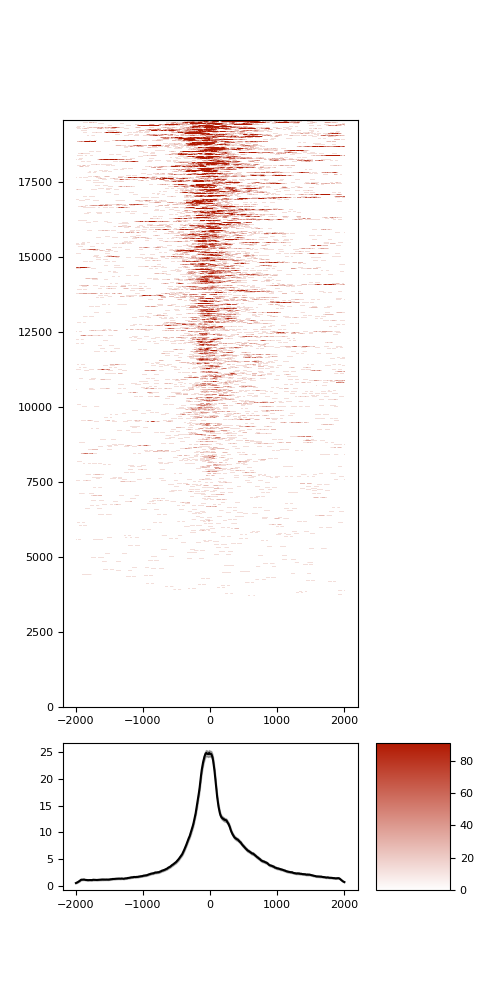
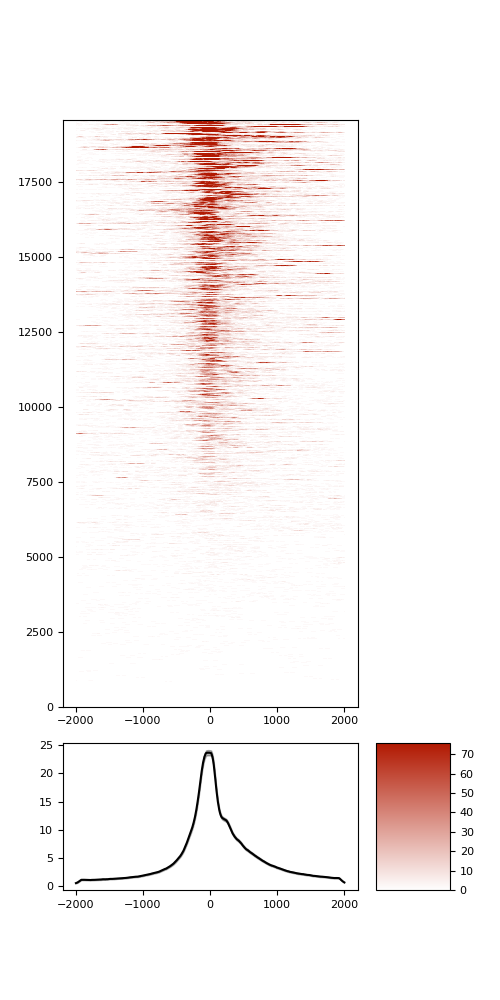
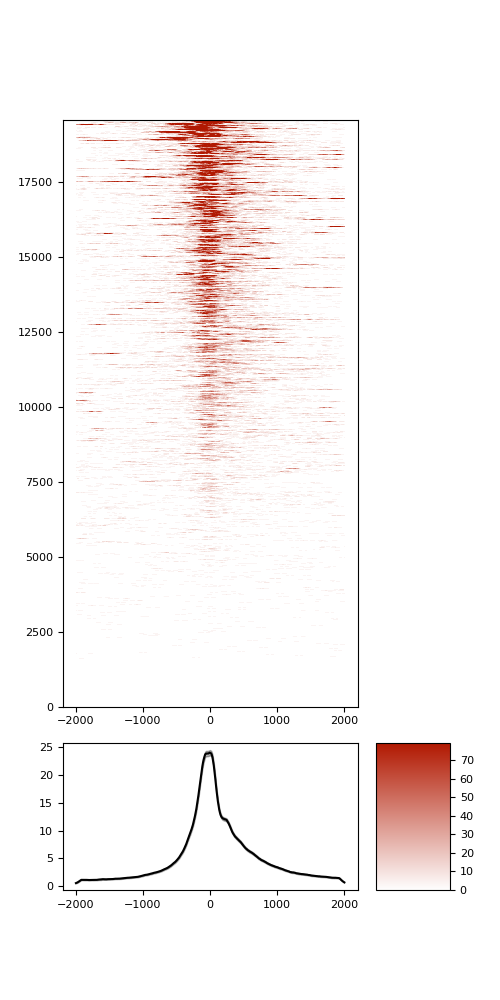
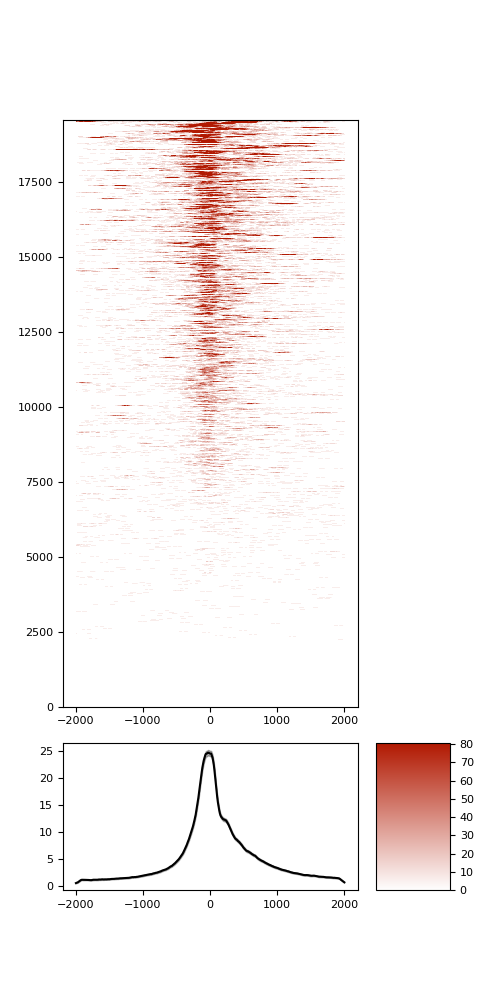
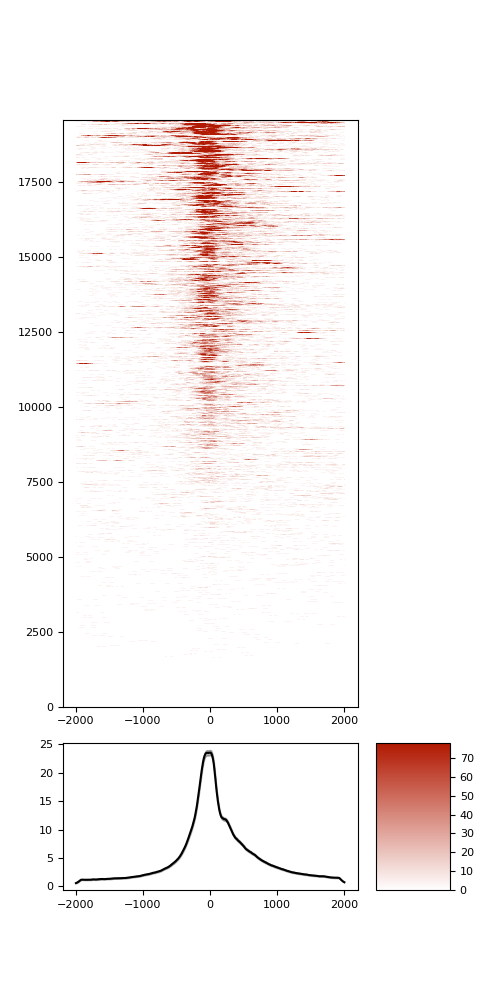
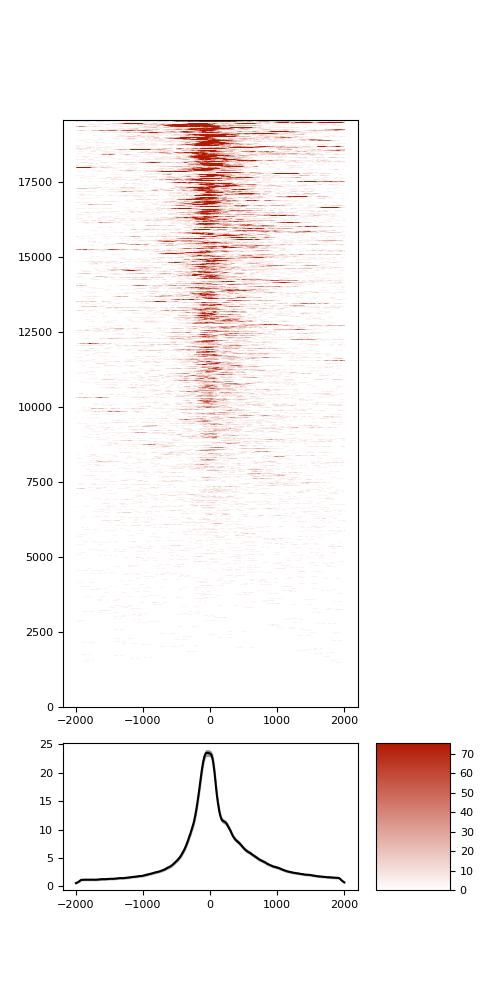
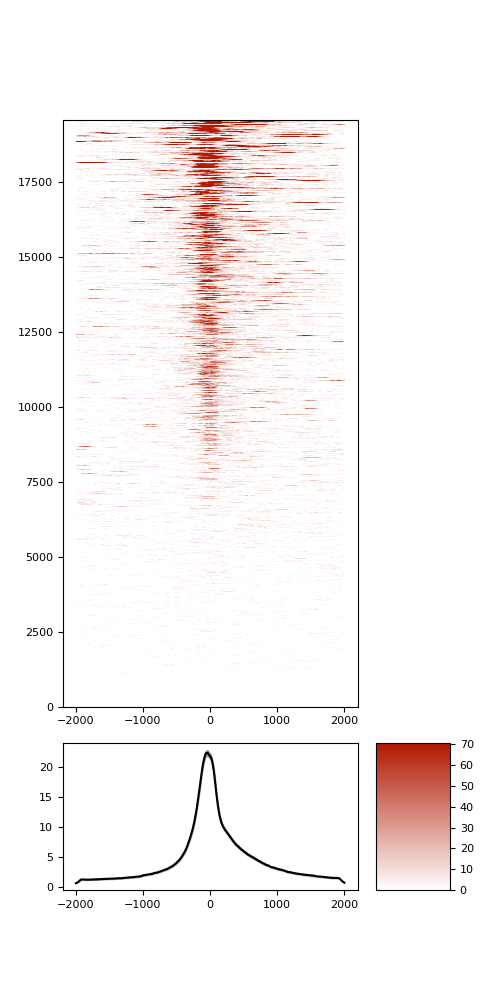
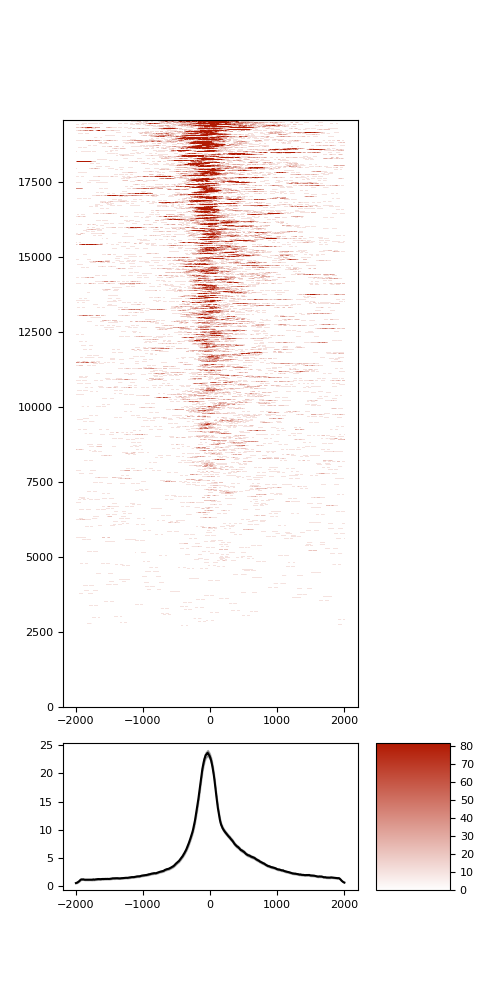
TSS enrichment
TSS enrichement plots
Average TSS enrichement compare
Max TSS enrichement compare
FROT (Fraction of reads overlap TSS)
Insert size & GC bias
Insert size distribution
GC bias in final bam
Peaks
Peak basics
Raw peak numbers
FRiP (Fraction of reads in Peak region)
Peak advanced
Correlation matrix & Peak Intensity Scatter
PCA
LibQC table
Tracks & Download
Batch download
click 'Download file list' link bellow to download a 'files.txt' that contains the list of urls to files in this report. Then run the script bellow on the terminal
Download file listxargs -n 1 curl -0 -L < files.txt